Chapter 13 The Philippines is similar
Philippines earthquakes are published by PHIVOLCS, and the web pages can be read directly into R. The approach used to gather this data automatically followed links on the page to capture the relevant information.
For the same period as the New Zealand Data (from September 2011 to September 2016), in earthquakes data from the Philippines there are 8054 events of depth greater than 0 and magnitude greater than 0.
| feature | value |
|---|---|
| Earliest (UTC) | 2011-09-01 |
| Latest (UTC) | 2016-08-29 13:30:00 |
| Northernmost | 21.47 |
| Southernmost | 2.2 |
| Westmost | 116.99 |
| Eastmost | 128.68 |
| Percent < Mag 3 | 40.81 |
| total entries | 8054 |
| nighttime quakes | 4959 |
Of the 8054 in the data, 4959 occurred at night, a proportion of 0.6157. A seven sigma confidence interval for the proportion of earthquakes occurring at night is 0.5773 to 0.6532, so I can be confident this is a non-random difference.
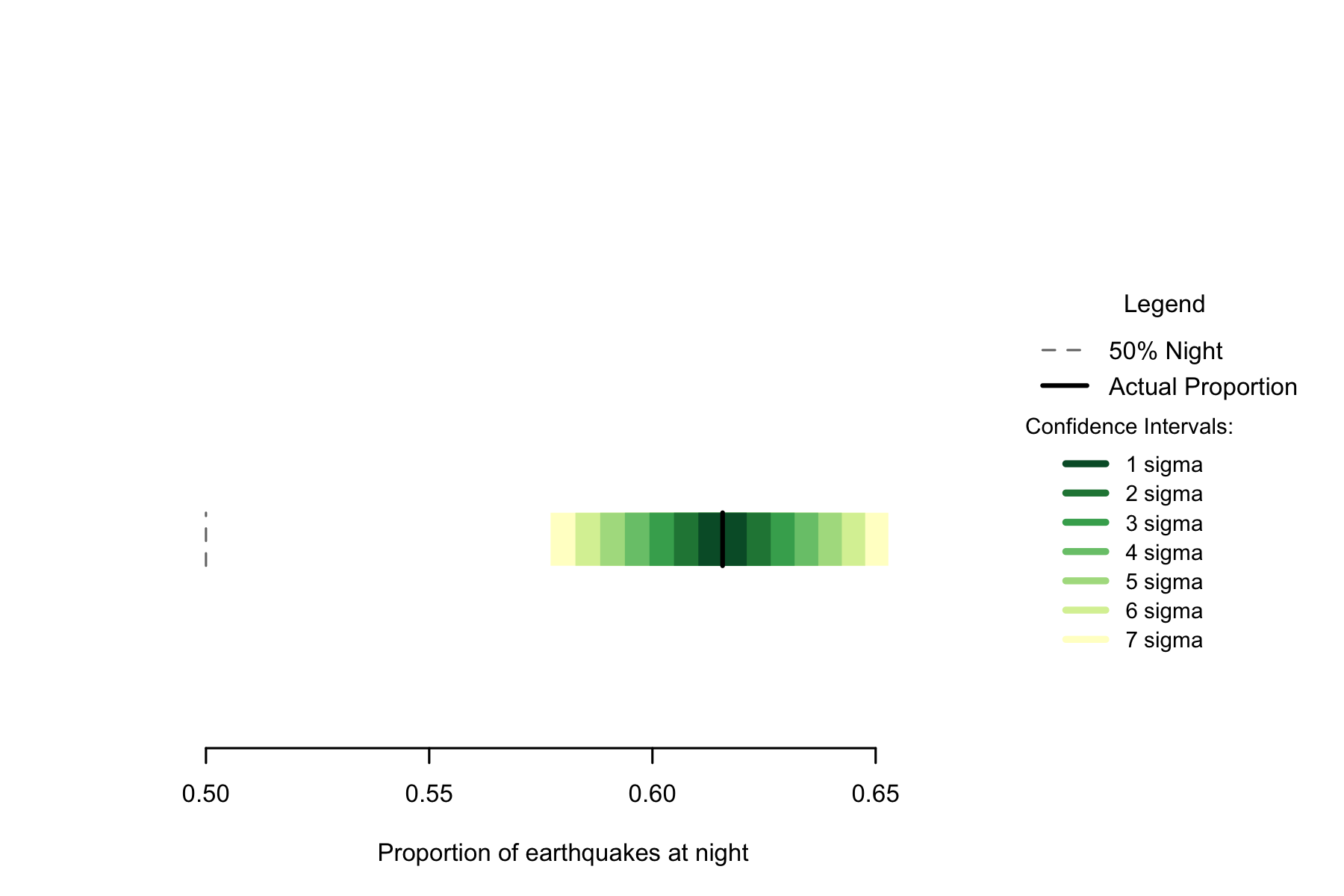
Figure 7.1: Proportion of earthquakes at night: Philippines. n=8054
The Philippines has the highest rate of nighttime earthquakes in a data set analysed in this study, so is clearly not 50% despite the small sample size.
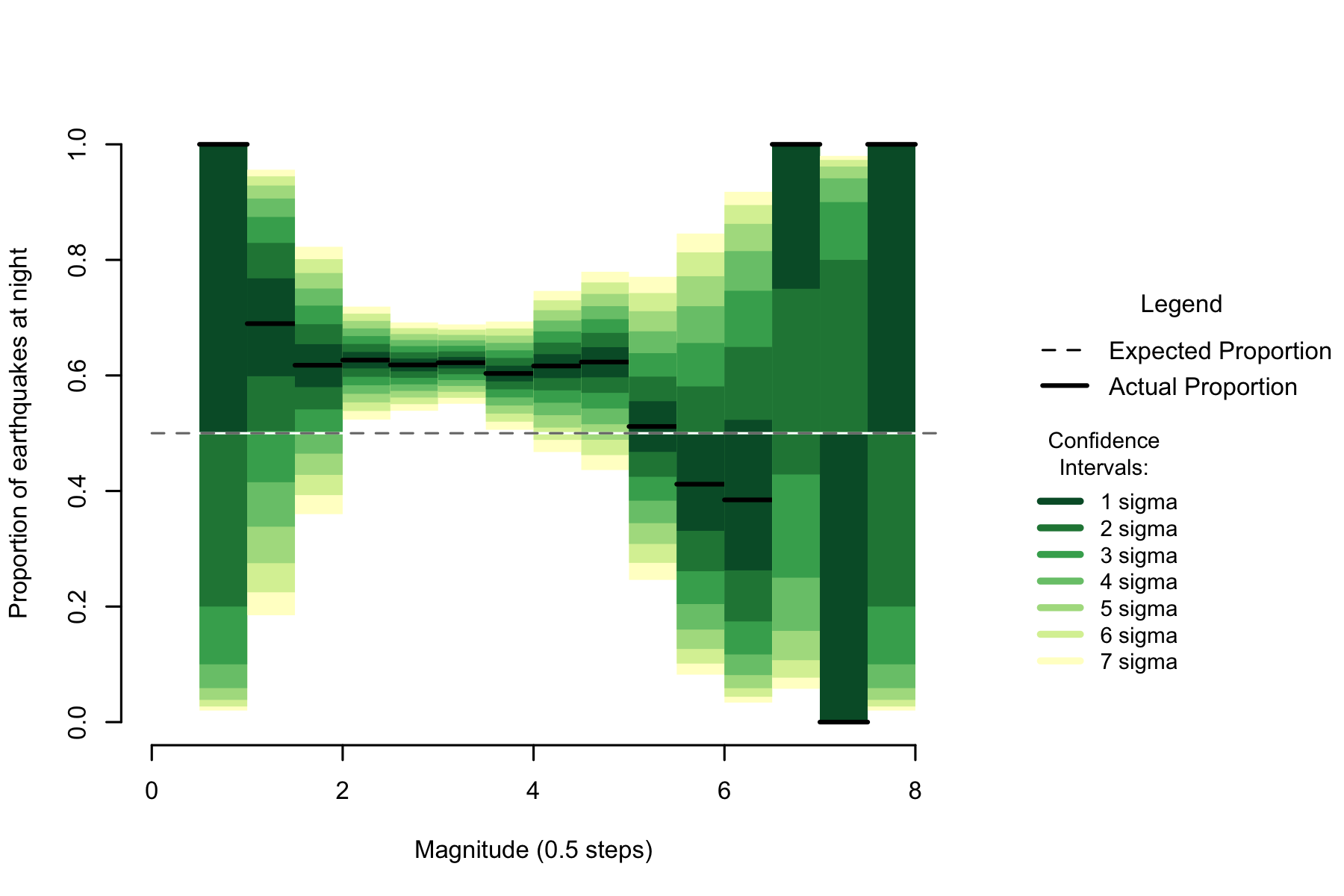
Figure 2.4: Proportion of night earthquakes by magnitude, Philippines. n=8054
Unusually, while the earthquakes from the Philippines show high night proportions at low magnitudes like other data sets, that rate does not unequivocally drop before the sample sizes shrink and the confidence intervals expand. While the Philippines has many fewer small earthquakes than previous data sets, the high night proportions effect extends to far higher magnitudes than in other regions data sets.
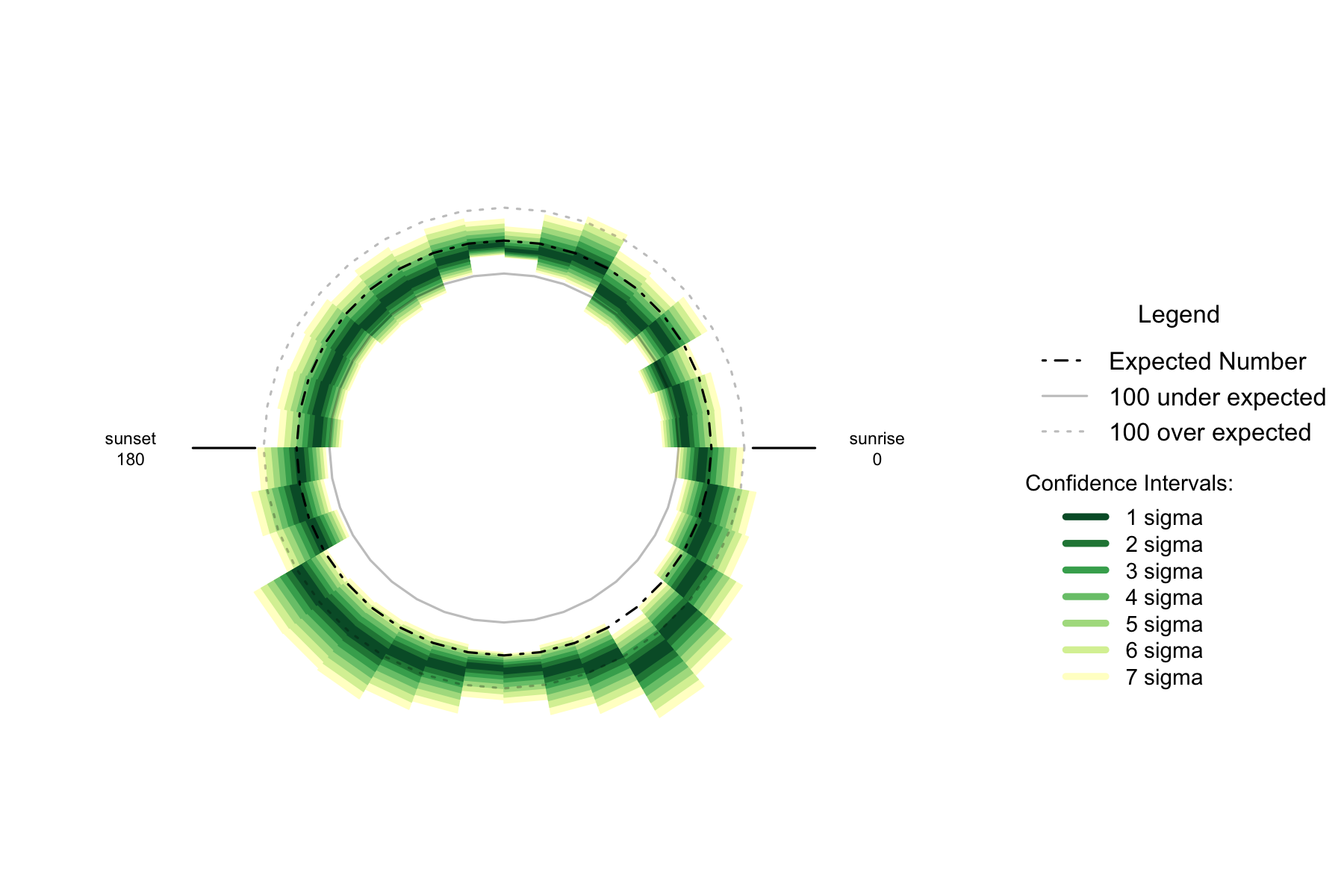
Figure 7.3: Over- and under- supply of earthquakes by angle of the sun (10 degree steps). , Philippines. n=8054
Night earthquakes show a general discontinuity to day, with a raised incidence (relative to day) of earthquakes through the entire night. There is still a peak at night when the sun is in the east, but not until 60 degrees below the horizon.
13.1 Formal Statement
Earthquakes in the Philippines show a similar pattern to New Zealand, displaying an oversupply of earthquakes at night that is not the result of chance. Despite a lack of small earthquakes, there is a clear bias towards night among small to medium magnitude earthquakes.
13.2 Links
1 - PHIVOLCS: http://www.phivolcs.dost.gov.ph/html/update_SOEPD/EQLatest.htm
13.3 Chapter Code
## ----setup, include=FALSE------------------------------------------------
knitr::opts_chunk$set(echo = FALSE)
## ----c10_001, warnings=FALSE, errors=FALSE, message=FALSE----------------
library(geosphere)
library(lubridate, quietly=TRUE)
library(dplyr)
library(binom)
library(ggplot2)
library(maps)
library(mapdata)
library(parallel)
library(readr)
library(plotrix)
library(tidyr)
library(maptools)
library(rvest)
Sys.setenv(TZ = "UTC")
## ----warnings=FALSE, errors=FALSE, message=FALSE-------------------------
if(!dir.exists("../othereqdata")){
dir.create("../othereqdata")
}
if(!file.exists("../othereqdata/eq_philippines_raw.RData")){
pg <- read_html("http://www.phivolcs.dost.gov.ph/html/update_SOEPD/EQLatest.html")
lnks <- pg %>% html_nodes("a") %>% html_attr("href") %>%
grep(pattern="(EQLatest-Monthly/)(2016|2015|2014|2013|2012|2011)", value=TRUE)
lnks <- paste("http://www.phivolcs.dost.gov.ph/html/update_SOEPD/", lnks, sep="")
assemble_eq <- function(x){
h <- html_text(read_html(x))
obs <- unlist(strsplit(gsub("([0-9]+) (Jan|Feb|Mar|Apr|May|Jun|Jul|Aug|Sep|Oct|Nov|Dec)",
"~\\1 \\2",h), "~" ))
obs <- obs[2:length(obs)]
obs <- gsub("(AM|PM)","\\1~",obs)
obs <- gsub("_+","", obs)
obs <- gsub("[[:space:]]+"," ", obs)
obs <- gsub("~ ","~",obs)
obsdf <- data.frame(obs) %>%
separate(obs, into=c("plpdate", "everything_else"), sep="~", extra="merge") %>%
separate(everything_else,
into=c("latitude", "longitude", "depth", "magnitude", "location"),
sep=" ", extra="merge") %>%
mutate(time_UTC = as.POSIXct(plpdate, format="%d %B %Y - %I:%M %p", tz="UTC") -
hours(8),
latitude = as.numeric(latitude),
longitude = as.numeric(longitude),
depth = as.numeric(depth),
magnitude = as.numeric(magnitude))
obsdf <- obsdf[!is.na(obsdf$latitude),]
return(obsdf)
}
eq_list <- lapply(lnks, assemble_eq)
eq_philippines <- bind_rows(eq_list)
eq_national <- eq_philippines %>%
filter(depth > 0 & magnitude >= 0 &
time_UTC >=
as.POSIXct("2011-09-01T00:00:00", format="%Y-%m-%dT%H:%M:%S", tz="UTC") &
time_UTC < as.POSIXct("2016-09-01T00:00:00",
format="%Y-%m-%dT%H:%M:%S", tz="UTC")) %>%
distinct() %>% arrange(time_UTC)
#one longitude was missing the decimal sign
eq_philippines$longitude[eq_philippines$longitude > 9000] <-
eq_philippines$longitude[eq_philippines$longitude > 9000] /100
rm(eq_philippines)
save(eq_national, file="../othereqdata/eq_philippines_raw.RData")
}
## ------------------------------------------------------------------------
if(!file.exists("../othereqdata/eq_philippines_processed.RData")){
load("../othereqdata/eq_philippines_raw.RData")
southmost <- min(eq_national$latitude)
westmost <- min(eq_national$longitude)
eq_national <- eq_national %>% filter(
magnitude > 0, depth > 0) %>% rowwise() %>% mutate(
eq_gridpoint_y = round(
distVincentyEllipsoid(c(longitude, southmost),c(longitude,latitude)) /50000,0),
eq_gridpoint_x = round(
distVincentyEllipsoid(c(westmost, latitude), c(longitude,latitude)) /50000,0),
eq_roundedlat = destPoint(
p=c(longitude, southmost), b=0, d=eq_gridpoint_y*50000)[2],
eq_roundedlong = destPoint(
p=c(westmost, eq_roundedlat), b=90, d=eq_gridpoint_x*50000)[1]) %>% ungroup()
# use maptools to calculate solar angles
sun_angles <- solarpos(
matrix(c(eq_national$longitude, eq_national$latitude), ncol=2), eq_national$time_UTC)
colnames(sun_angles) <- c("eq_compass", "eq_vertical")
eq_national <- cbind(eq_national,sun_angles)
eq_national$eq_is_night <- eq_national$eq_vertical < 0
# calculate 360 degree as well as vertical
eq_national <- eq_national %>%
mutate(eq_angle_360 = eq_vertical,
eq_angle_360 = ifelse(eq_compass > 180, 180 - eq_angle_360, eq_angle_360),
eq_angle_360 = ifelse(eq_vertical < 0 & eq_compass <= 180,
360 + eq_angle_360, eq_angle_360),
eq_angle_by_10 = floor(eq_angle_360 /10) * 10)
save(eq_national, file="../othereqdata/eq_philippines_processed.RData")
}
## ------------------------------------------------------------------------
if(!file.exists("../othereqdata/eq_philippines_expected.RData")){
load("../othereqdata/eq_philippines_processed.RData")
lat_range <- unique(eq_national$eq_roundedlat)
long_med <- median(eq_national$eq_roundedlong)
# 1 minute intervals for a full solar year
time1 <- ymd_hms("2015-01-01 00:00:00")
time2 <- ymd_hms("2015-12-31 23:59:00")
time_sq <- seq.POSIXt(from=time1, to=time2, by="min")
calc_angs <- function(x, longinput, timeinput){
library(dplyr)
sun_angles <- maptools::solarpos(matrix(c(longinput, x), ncol=2), timeinput)
colnames(sun_angles) <- c("eq_compass", "eq_vertical")
# calculate 360 degree as well as vertical
site_summary <- as.data.frame(sun_angles) %>%
mutate(eq_angle_360 = eq_vertical,
eq_angle_360 = ifelse(eq_compass > 180, 180 - eq_angle_360, eq_angle_360),
eq_angle_360 = ifelse(
eq_vertical < 0 & eq_compass <= 180, 360 + eq_angle_360, eq_angle_360),
eq_angle_by_10 = floor(eq_angle_360 /10) * 10) %>%
group_by(eq_angle_by_10) %>% summarise(total= n())
site_summary$lat <- x
return(site_summary)
}
###
# Calculate the number of cores
no_cores <- detectCores() - 1
# Initiate cluster
cl <- makeCluster(no_cores)
clusterExport(cl, varlist=c("lat_range", "long_med", "time_sq", "calc_angs"))
list_angs <- parLapply(cl, lat_range,
function(x){
calc_angs(x=x, longinput=long_med, timeinput=time_sq)})
stopCluster(cl)
###
library(tidyr)
anglong <- bind_rows(list_angs)
angwide <- spread(anglong, key=eq_angle_by_10,value=total)
rm(anglong, list_angs, time_sq)
save(angwide, file="../othereqdata/eq_philippines_expected.RData")
}
## ------------------------------------------------------------------------
load("../othereqdata/eq_philippines_processed.RData")
load("../othereqdata/eq_philippines_expected.RData")
eq_night = sum(eq_national$eq_is_night)
eq_total = nrow(eq_national)
bands <- rev(c('#ffffcc','#d9f0a3','#addd8e','#78c679','#41ab5d','#238443','#005a32'))
sigmas <- c(0.682689492137086,
0.954499736103642,
0.997300203936740,
0.999936657516334,
0.999999426696856,
0.999999998026825,
0.999999999997440)
lbls <- c(
"1 sigma", "2 sigma",
"3 sigma", "4 sigma",
"5 sigma", "6 sigma",
"7 sigma")
typs <- c(1,1,1,1,1,1,1)
weights <- c(3,3,3,3,3,3,3)
old_par=par()
## ------------------------------------------------------------------------
bt <- binom.test(eq_night ,eq_total, conf.level= .999999999997440)
## ------------------------------------------------------------------------
feature <- c("Earliest (UTC)", "Latest (UTC)",
"Northernmost", "Southernmost",
"Westmost", "Eastmost",
"Percent < Mag 3", "total entries",
"nighttime quakes")
value <- c(as.character(min(eq_national$time_UTC)),
as.character(max(eq_national$time_UTC)),
as.character(max(eq_national$latitude)),
as.character(min(eq_national$latitude)),
as.character(min(eq_national$longitude)),
as.character(max(eq_national$longitude)),
as.character(round(100*sum(eq_national$magnitude < 3)/eq_total,2)),
as.character(eq_total),
as.character(eq_night))
data.frame(feature,value) %>% knitr::kable(caption="Data description")
## ---- fig.cap="Proportion of earthquakes at night: Philippines. n=8054"----
### making the basic proportion graph
eq_night = sum(eq_national$eq_is_night)
eq_total = nrow(eq_national)
bands <- rev(c('#ffffcc','#d9f0a3','#addd8e','#78c679','#41ab5d','#238443','#005a32'))
sigmas <- c(0.682689492137086,
0.954499736103642,
0.997300203936740,
0.999936657516334,
0.999999426696856,
0.999999998026825,
0.999999999997440)
lbls <- c(
"1 sigma", "2 sigma",
"3 sigma", "4 sigma",
"5 sigma", "6 sigma",
"7 sigma")
typs <- c(1,1,1,1,1,1,1)
weights <- c(3,3,3,3,3,3,3)
old_par=par()
conf_steps <- function(x, sigmas=sigmas, night=eq_night, total=eq_total){
ci_lower <- binom.confint(night, total, method=c("wilson"), conf.level = sigmas[x])[1,5]
ci_upper <- binom.confint(night, total, method=c("wilson"), conf.level = sigmas[x])[1,6]
ci_data <- data.frame(step = x, ci_lower, ci_upper)
}
ci_spacing <- lapply(7:1, conf_steps, sigmas=sigmas, night=eq_night, total=eq_total)
ci_steps <- bind_rows(ci_spacing)
layout(matrix(c(1,1,1,2), ncol=4))
par(mar=c(5,6,4,2))
plot(c(min(0.5,floor(100*ci_steps[1,2])/100), max(0.5,ceiling(100*ci_steps[1,3])/100)),
y=c(-3,8), type="n", bty="n", yaxt="n", ylab="",
xlab="Proportion of earthquakes at night")
a <- a <- apply(ci_steps, 1, function(x){
polygon(c(x[2], x[3], x[3], x[2]), c(0, 0, 1, 1), col=bands[x[1]], border=NA)})
lines(c(.5,.5), c(0,1), col="#FFFFFF")
lines(c(.5,.5), c(0,1), lty=2, col="#777777")
lines(c(eq_night/eq_total,eq_night/eq_total), c(0,1), lwd=2)
par(mar=c(0,0,0,0))
plot(x=c(0,10), y=c(0,10), type="n", bty="n", axes=FALSE)
legend(0,5.5, legend=lbls, lty=typs, lwd=weights, col=bands, bty="n", xjust=0,
title="Confidence Intervals:", y.intersp=1.1, cex=0.9)
lbls2=c("50% Night", "Actual Proportion")
typs2=c(2,1)
weights2=c(1,2)
cls2=c("#777777","#000000")
legend(0,7, legend=lbls2, lty=typs2, lwd=weights2, col=cls2, bty="n", xjust=0,
title="Legend", y.intersp=1.2)
par(mar=old_par$mar)
par(mfrow=c(1,1))
## ---- fig.cap="Proportion of night earthquakes by magnitude, Philippines. n=8054"----
old_par=par()
grf <- eq_national %>% mutate(floored_mag = floor(magnitude*2)/2) %>%
group_by(floored_mag) %>% summarise(successes = sum(eq_is_night), trials=n())
poly_conf_int <- function(success, trials, aa, stepsize, sigma, colr){
ci <- binom.confint(success, trials, method=c("wilson"), conf.level = sigma)
lower <- ci[1,5]
upper <- ci[1,6]
a <- polygon(x=c(aa,aa+stepsize,aa+stepsize,aa), y=c(upper,upper,lower,lower),
col=colr, border=NA)
}
plot7sig <- function(success, trials, aa, stepsize){
library(binom)
#bands <- c('#ffffb2','#fed976','#feb24c','#fd8d3c','#fc4e2a','#e31a1c','#b10026')
bands <- rev(c('#ffffcc','#d9f0a3','#addd8e','#78c679','#41ab5d','#238443','#005a32'))
sigmas <- c(0.682689492137086,
0.954499736103642,
0.997300203936740,
0.999936657516334,
0.999999426696856,
0.999999998026825,
0.999999999997440)
sapply(7:1, function(x){
poly_conf_int(success, trials, aa, stepsize, sigmas[x], bands[x])})
a <- lines(c(aa, aa + stepsize), c(success/trials, success/trials), lwd=2)
}
lbls <- c(
"1 sigma", "2 sigma",
"3 sigma", "4 sigma",
"5 sigma", "6 sigma",
"7 sigma")
typs <- c(1,1,1,1,1,1,1)
weights <- c(3,3,3,3,3,3,3)
clrs <- rev(c('#ffffcc','#d9f0a3','#addd8e','#78c679','#41ab5d','#238443','#005a32'))
#clrs <- c('#ffffb2','#fed976','#feb24c','#fd8d3c','#fc4e2a','#e31a1c','#b10026')
layout(matrix(c(1,1,1,2), ncol=4))
plot(x=c(0,max(grf$floored_mag)+0.5), y=c(0,1), type="n", bty="n",
xlab="Magnitude (0.5 steps)", ylab="Proportion of earthquakes at night")
a <- apply(grf,1,function(x){plot7sig(x[2],x[3],x[1],0.5)})
lines(c(0,10), c(.5,.5), col="#FFFFFF")
lines(c(0,10), c(.5,.5), lty=2, col="#777777")
par(mar=c(0,0,0,0))
plot(x=c(0,10), y=c(0,10), type="n", bty="n", axes=FALSE)
legend(0,5, legend=lbls, lty=typs, lwd=weights, col=clrs, bty="n", xjust=0,
title="Confidence
Intervals:", cex=0.9)
lbls=c("Expected Proportion", "Actual Proportion")
typs=c(2,1)
weights=c(1,2)
legend(0,7, legend=lbls, lty=typs, lwd=weights, bty="n", xjust=0,
title="Legend", y.intersp=1.2)
par(mar=old_par$mar)
par(mfrow=c(1,1))
## ------------------------------------------------------------------------
by_angle <- eq_national %>%
group_by(eq_angle_by_10) %>% summarise(total= n()) %>%
mutate(daynight=ifelse(eq_angle_by_10 < 180, "day", "night"))
merged <- merge(eq_national, angwide, by.x="eq_roundedlat", by.y="lat")
agg_expected <- merged %>% select(`0`:`350`) %>% colSums(na.rm=TRUE)
expected_prop <- agg_expected / sum(agg_expected)
expected <- data.frame(eq_angle_by_10 = as.numeric(names(expected_prop)),
expected_prop = as.numeric(expected_prop))
expected$expected_number = expected_prop * eq_total
act_exp <- merge(expected, by_angle, by="eq_angle_by_10", all.x=TRUE)
act_exp$total[is.na(act_exp$total)] <- 0
act_exp$daynight <- NULL
act_exp$act_prop <- act_exp$total / sum(act_exp$total)
ci_brackets <- act_exp %>% ungroup() %>% mutate(grand_total=sum(total)) %>%
rowwise() %>% mutate(
ci_lower_7 = binom.confint(total, grand_total, method=c("wilson"),
conf.level = sigmas[7])[1,5] * grand_total,
ci_upper_7 = binom.confint(total, grand_total, method=c("wilson"),
conf.level = sigmas[7])[1,6] * grand_total,
ci_lower_6 = binom.confint(total, grand_total, method=c("wilson"),
conf.level = sigmas[6])[1,5] * grand_total,
ci_upper_6 = binom.confint(total, grand_total, method=c("wilson"),
conf.level = sigmas[6])[1,6] * grand_total,
ci_lower_5 = binom.confint(total, grand_total, method=c("wilson"),
conf.level = sigmas[5])[1,5] * grand_total,
ci_upper_5 = binom.confint(total, grand_total, method=c("wilson"),
conf.level = sigmas[5])[1,6] * grand_total,
ci_lower_4 = binom.confint(total, grand_total, method=c("wilson"),
conf.level = sigmas[4])[1,5] * grand_total,
ci_upper_4 = binom.confint(total, grand_total, method=c("wilson"),
conf.level = sigmas[4])[1,6] * grand_total,
ci_lower_3 = binom.confint(total, grand_total, method=c("wilson"),
conf.level = sigmas[3])[1,5] * grand_total,
ci_upper_3 = binom.confint(total, grand_total, method=c("wilson"),
conf.level = sigmas[3])[1,6] * grand_total,
ci_lower_2 = binom.confint(total, grand_total, method=c("wilson"),
conf.level = sigmas[2])[1,5] * grand_total,
ci_upper_2 = binom.confint(total, grand_total, method=c("wilson"),
conf.level = sigmas[2])[1,6] * grand_total,
ci_lower_1 = binom.confint(total, grand_total, method=c("wilson"),
conf.level = sigmas[1])[1,5] * grand_total,
ci_upper_1 = binom.confint(total, grand_total, method=c("wilson"),
conf.level = sigmas[1])[1,6] * grand_total)
norm_ci <- ci_brackets
for (i in c(4,7:20)){
norm_ci[,i] <- ci_brackets[,i] - ci_brackets[,3]
}
circlesize=100
## ---- fig.cap="Over- and under- supply of earthquakes by angle of the sun
## (10 degree steps). , Philippines. n=8054"----
norm_ci$border = 2
# need to double entries with a displacement of 10
# to make each side of the item on the graph
norm_ci2 <- norm_ci
norm_ci2$eq_angle_by_10 <- norm_ci2$eq_angle_by_10 + 10
norm_ci2$border = 1
graphdata <- bind_rows(norm_ci,norm_ci2) %>% arrange(eq_angle_by_10,border)
#### plot graph
bands <- rev(c('#ffffcc','#d9f0a3','#addd8e','#78c679','#41ab5d','#238443','#005a32'))
old_par=par()
layout(matrix(c(1,1,1,2), ncol=4))
# overall limits
limits=2 * max(abs(c(graphdata$ci_lower_7, graphdata$ci_upper_7)))
# plot upper confidence 7 interval using plotrix
polar.plot(graphdata$ci_upper_7, polar.pos=graphdata$eq_angle_by_10,
radial.lim=c(-1*limits,limits),
labels = "", main=NULL,lwd=0.5, rp.type="p",
show.grid.labels=FALSE, show.grid=FALSE, mar=c(0,0,0,0),
grid.col=bands[7], line.col=bands[7], poly.col=bands[7])
# plot upper 6 confidence interval
plot_ci_round <- function(upper_bound,x){
polar.plot(upper_bound, polar.pos=graphdata$eq_angle_by_10, add=TRUE,
radial.lim=c(-1*limits,limits),
line.col=bands[x], lwd=0.5, rp.type="p", poly.col=bands[x])
}
plot_ci_round(graphdata$ci_upper_6, 6)
plot_ci_round(graphdata$ci_upper_5, 5)
plot_ci_round(graphdata$ci_upper_4, 4)
plot_ci_round(graphdata$ci_upper_3, 3)
plot_ci_round(graphdata$ci_upper_2, 2)
plot_ci_round(graphdata$ci_upper_1, 1)
plot_ci_round(graphdata$ci_lower_1, 2)
plot_ci_round(graphdata$ci_lower_2, 3)
plot_ci_round(graphdata$ci_lower_3, 4)
plot_ci_round(graphdata$ci_lower_4, 5)
plot_ci_round(graphdata$ci_lower_5, 6)
plot_ci_round(graphdata$ci_lower_6, 7)
polar.plot(graphdata$ci_lower_7, polar.pos=graphdata$eq_angle_by_10,
add=TRUE, radial.lim=c(-1*limits,limits),
line.col="white", lwd=0.5, rp.type="p", poly.col="white")
# plot expected guide line
polar.plot(rep(0,nrow(graphdata)), polar.pos=graphdata$eq_angle_by_10, add=TRUE,
radial.lim=c(-1*limits,limits),
rp.type="p", lty=4)
# plot 500 less than expected guide line
polar.plot(rep(-1 * circlesize,nrow(graphdata)), polar.pos=graphdata$eq_angle_by_10,
add=TRUE,radial.lim=c(-1*limits,limits),
rp.type="p", lty=1, line.col="#00000044")
# plot 500 more than expected guide line
polar.plot(rep(circlesize,nrow(graphdata)), polar.pos=graphdata$eq_angle_by_10,
add=TRUE,radial.lim=c(-1*limits,limits),
rp.type="p", lty=3, line.col="#00000044")
lines(c(-1.5,-1.2)*limits, c(0,0))
lines(c(1.5,1.2)*limits, c(0,0))
text(-1.8*limits,0, label="sunset
180", cex=0.7)
text(1.8*limits,0, label="sunrise
0", cex=0.7)
par(mar=c(0,0,0,0))
plot(x=c(0,10), y=c(0,10), type="n", bty="n", axes=FALSE, xlab="")
lbls <- c(
"1 sigma", "2 sigma",
"3 sigma", "4 sigma",
"5 sigma", "6 sigma",
"7 sigma")
typs <- c(1,1,1,1,1,1,1)
weights <- c(3,3,3,3,3,3,3)
clrs <- rev(c('#ffffcc','#d9f0a3','#addd8e','#78c679','#41ab5d','#238443','#005a32'))
legend(0,4.5, legend=lbls, lty=typs, lwd=weights, col=clrs, bty="n", xjust=0,
title="Confidence Intervals:", cex=0.9)
lbls2=c("Expected Number", paste(circlesize,"under expected"),
paste(circlesize,"over expected"))
typs2=c(4,1,3)
weights2=c(1,1,1)
clrs2=c("#000000","#00000044","#00000044")
legend(0,10, legend=lbls2, lty=typs2, lwd=weights2, bty="n", xjust=0,
title="Legend", y.intersp=1.2, col=clrs2)
par(mar=old_par$mar)
par(mfrow=c(1,1))