Chapter 16 Australia is similar
Australian earthquake data is publicly made available from Geoscience Australia. This data was obtained by searching the database for all Australia and download the result as csv, then reading the saved csv into R.
For the same period as the New Zealand Data (from September 2011 to September 2016), in earthquakes data from the Australia there are 3182 events of depth greater than 0 and magnitude greater than 0.
| feature | value |
|---|---|
| Earliest (UTC) | 2011-09-01 01:56:51 |
| Latest (UTC) | 2016-08-31 16:25:18 |
| Northernmost | -11.546 |
| Southernmost | -43.429 |
| Westmost | 111.085 |
| Eastmost | 154.521 |
| Percent < Mag 3 | 84.82 |
| total entries | 3182 |
| nighttime quakes | 1911 |
Of the 3182 in the data, 1911 occurred at night, a proportion of 0.6006. A seven sigma confidence interval for the proportion of earthquakes occurring at night is 0.5388 to 0.6602, so I can be confident this is a non-random difference. This very high rate is narrowly smaller than the rate observed in the Philippines.
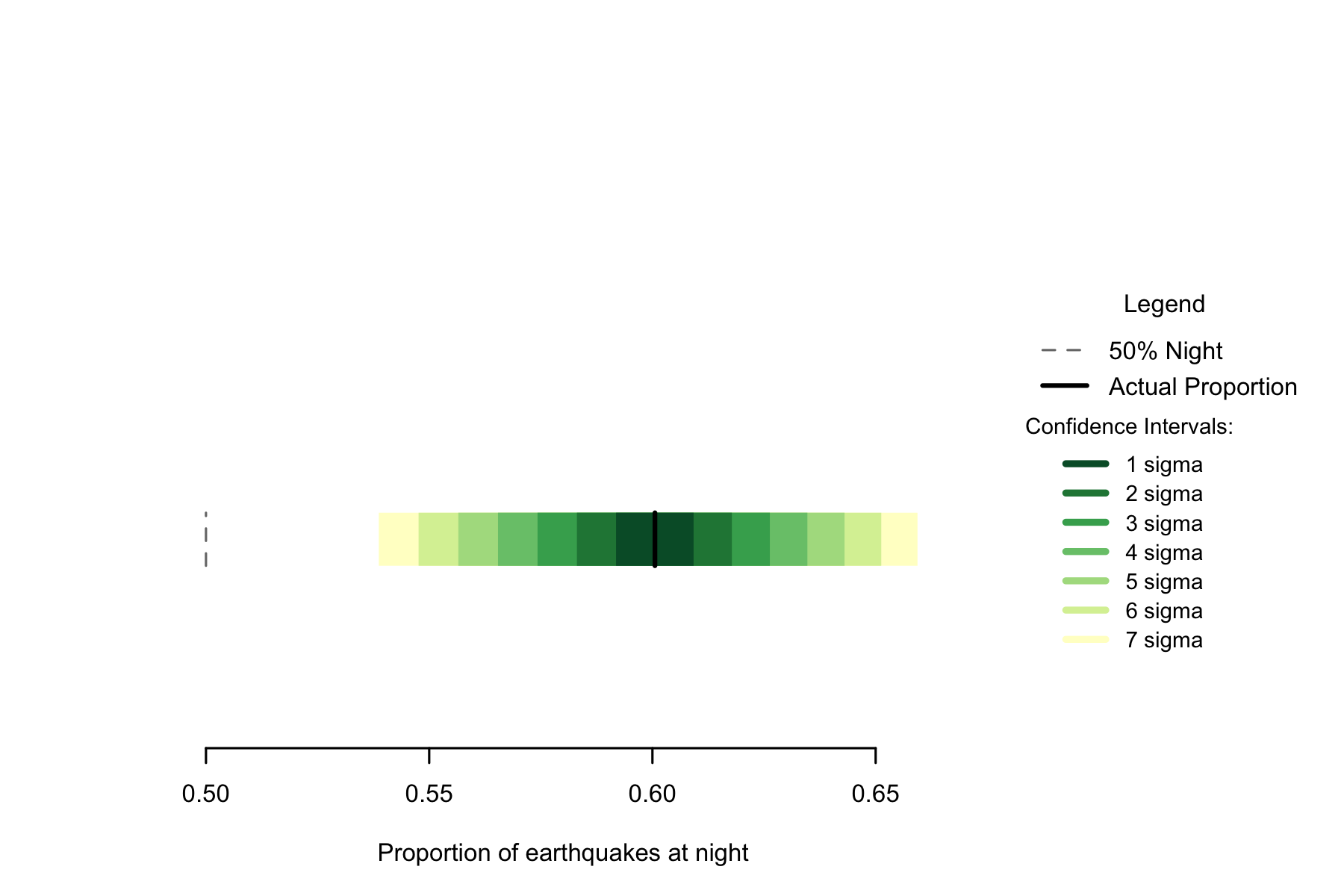
Figure 7.1: Proportion of earthquakes at night: Philippines. n=8054
Because of the strength of the effect in Australia, it is clearly not 50% despite the small sample size leading to large confidence intervals.
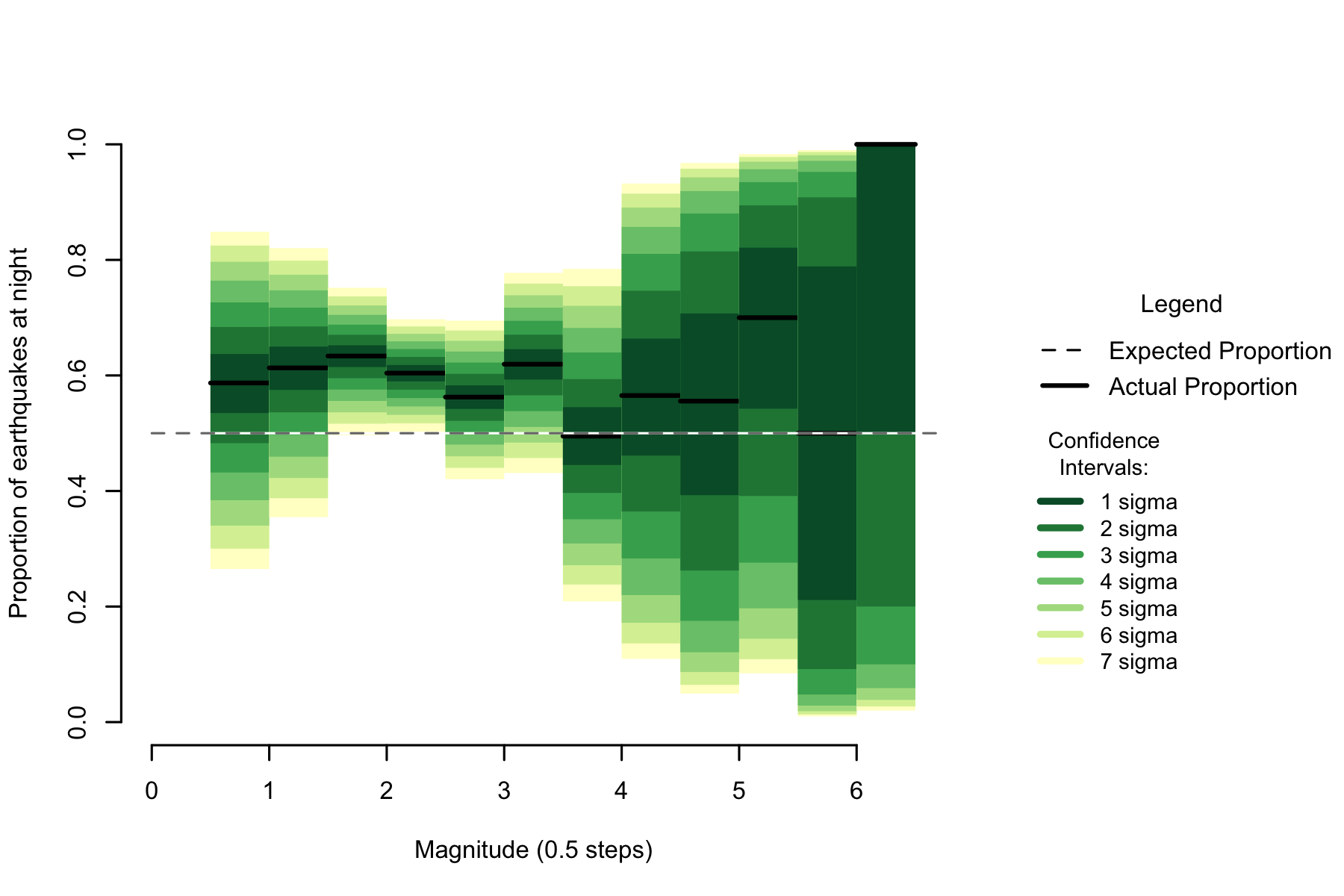
Figure 2.4: Proportion of night earthquakes by magnitude, Philippines. n=8054
Australia seems to show a clearly high rate of night quakes in the 1 to 2 magnitude range, but due to sample size it is hard to form an opinion on if that rate is holding until 3.5 or dropping towards 50%.
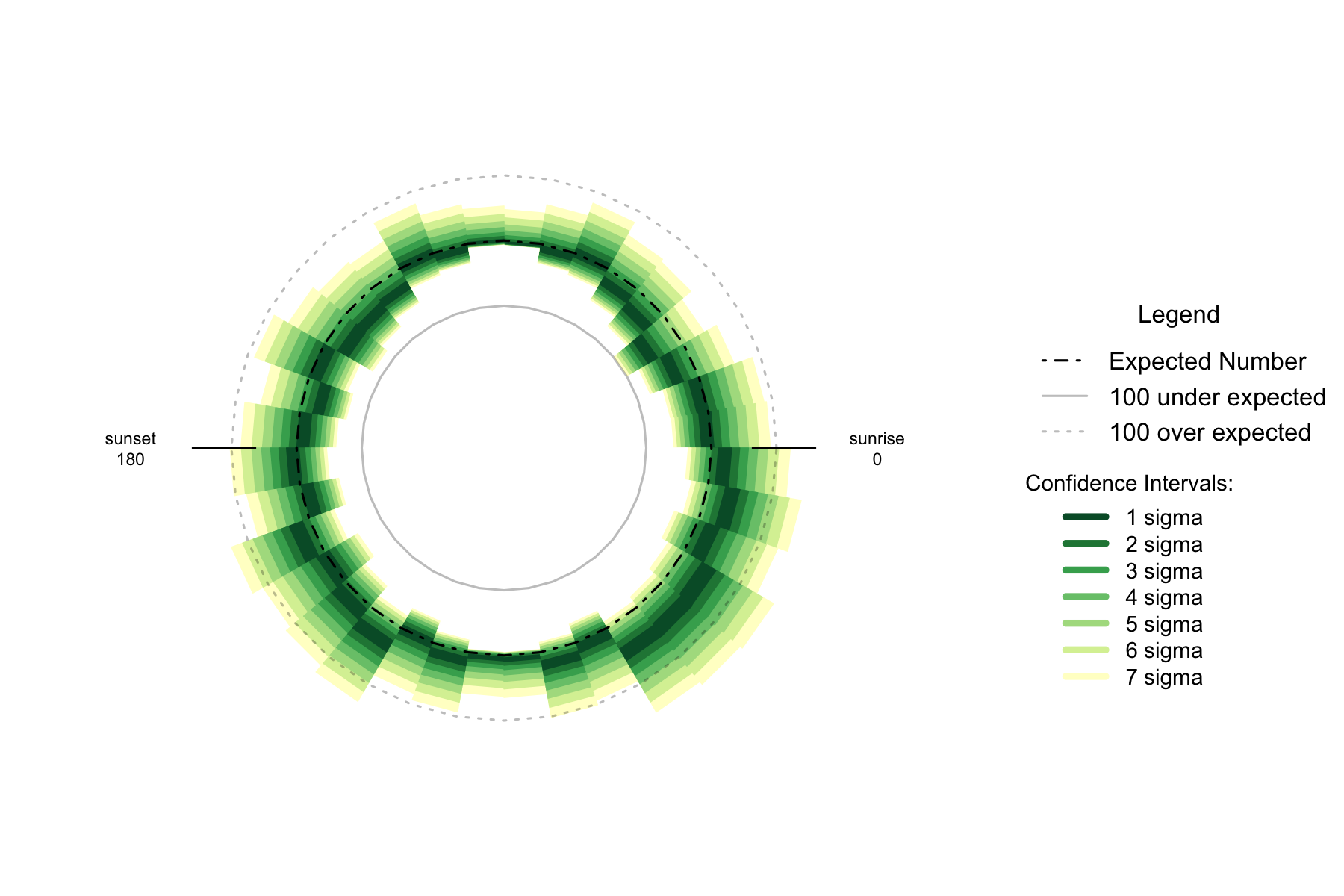
Figure 7.3: Over- and under- supply of earthquakes by angle of the sun (10 degree steps). Hawaii. n=10181
By the time the earthquakes are divided into 10 degree arcs of the sun, sample sizes are a little small to say much useful about the Australian data. When the sun is in the east below the horizon seems to consistently have more earthquakes than other times, but there are not enough sample to form more detailed conclusions.
16.1 Formal Statement
Earthquakes in the Australia show a similar pattern to New Zealand, displaying an oversupply of earthquakes at night that is not the result of chance, and clear evidence of smaller magnitude earthquakes having a high rate of nighttime occurrence. The sample size does not permit detailed conclusions about the pattern with respect to the angle of the sun at time of earthquake.
16.2 Links
1 - Geoscience Australia Earthquakes: http://www.ga.gov.au/earthquakes/searchQuake.do
16.3 Chapter Code
## ----setup, include=FALSE------------------------------------------------
knitr::opts_chunk$set(echo = FALSE)
## ----c10_001, warnings=FALSE, errors=FALSE, message=FALSE----------------
library(geosphere)
library(lubridate, quietly=TRUE)
library(dplyr)
library(binom)
library(ggplot2)
library(maps)
library(mapdata)
library(parallel)
library(readr)
library(plotrix)
library(tidyr)
library(maptools)
library(rvest)
Sys.setenv(TZ = "UTC")
## ----warnings=FALSE, errors=FALSE, message=FALSE-------------------------
if(!dir.exists("../othereqdata")){
dir.create("../othereqdata")
}
if(!file.exists("../othereqdata/eq_australia_raw.RData")){
eq_aus <- read.csv("../australia/eq_103_1492071234067.csv", stringsAsFactors = FALSE)
eq_aus$time_UTC <- ymd_hms(paste(eq_aus$UTC.Date, eq_aus$UTC.Time))
names(eq_aus)[c(1,6,7,9)] <- c("magnitude",
"latitude", "longitude", "depth")
eq_national <- eq_aus %>%
filter(magnitude >= 0 &
time_UTC >= as.POSIXct("2011-09-01T00:00:00",
format="%Y-%m-%dT%H:%M:%S", tz="UTC") &
time_UTC < as.POSIXct("2016-09-01T00:00:00",
format="%Y-%m-%dT%H:%M:%S", tz="UTC")) %>%
distinct() %>% arrange(time_UTC)
#one longitude was missing the decimal sign
rm(eq_aus)
save(eq_national, file="../othereqdata/eq_australia_raw.RData")
}
## ------------------------------------------------------------------------
if(!file.exists("../othereqdata/eq_australia_processed.RData")){
load("../othereqdata/eq_australia_raw.RData")
southmost <- min(eq_national$latitude)
westmost <- min(eq_national$longitude)
eq_national <- eq_national %>% filter(
magnitude > 0) %>% rowwise() %>% mutate(
eq_gridpoint_y = round(distVincentyEllipsoid(c(longitude, southmost),
c(longitude,latitude)) /50000,0),
eq_gridpoint_x = round(distVincentyEllipsoid(c(westmost, latitude),
c(longitude,latitude)) /50000,0),
eq_roundedlat = destPoint(p=c(longitude, southmost),
b=0, d=eq_gridpoint_y*50000)[2],
eq_roundedlong = destPoint(p=c(westmost, eq_roundedlat),
b=90, d=eq_gridpoint_x*50000)[1]) %>% ungroup()
# use maptools to calculate solar angles
sun_angles <- solarpos(matrix(c(eq_national$longitude, eq_national$latitude), ncol=2),
eq_national$time_UTC)
colnames(sun_angles) <- c("eq_compass", "eq_vertical")
eq_national <- cbind(eq_national,sun_angles)
eq_national$eq_is_night <- eq_national$eq_vertical < 0
# calculate 360 degree as well as vertical
eq_national <- eq_national %>%
mutate(eq_angle_360 = eq_vertical,
eq_angle_360 = ifelse(eq_compass > 180, 180 - eq_angle_360, eq_angle_360),
eq_angle_360 = ifelse(eq_vertical < 0 & eq_compass <= 180,
360 + eq_angle_360, eq_angle_360),
eq_angle_by_10 = floor(eq_angle_360 /10) * 10)
save(eq_national, file="../othereqdata/eq_australia_processed.RData")
}
## ------------------------------------------------------------------------
if(!file.exists("../othereqdata/eq_australia_expected.RData")){
load("../othereqdata/eq_australia_processed.RData")
lat_range <- unique(eq_national$eq_roundedlat)
long_med <- median(eq_national$eq_roundedlong)
# 1 minute intervals for a full solar year
time1 <- ymd_hms("2015-01-01 00:00:00")
time2 <- ymd_hms("2015-12-31 23:59:00")
time_sq <- seq.POSIXt(from=time1, to=time2, by="min")
calc_angs <- function(x, longinput, timeinput){
library(dplyr)
sun_angles <- maptools::solarpos(matrix(c(longinput, x), ncol=2), timeinput)
colnames(sun_angles) <- c("eq_compass", "eq_vertical")
# calculate 360 degree as well as vertical
site_summary <- as.data.frame(sun_angles) %>%
mutate(eq_angle_360 = eq_vertical,
eq_angle_360 = ifelse(eq_compass > 180, 180 - eq_angle_360, eq_angle_360),
eq_angle_360 = ifelse(
eq_vertical < 0 & eq_compass <= 180, 360 + eq_angle_360, eq_angle_360),
eq_angle_by_10 = floor(eq_angle_360 /10) * 10) %>%
group_by(eq_angle_by_10) %>% summarise(total= n())
site_summary$lat <- x
return(site_summary)
}
###
# Calculate the number of cores
no_cores <- detectCores() - 1
# Initiate cluster
cl <- makeCluster(no_cores)
clusterExport(cl, varlist=c("lat_range", "long_med", "time_sq", "calc_angs"))
list_angs <- parLapply(cl, lat_range,
function(x){
calc_angs(x=x, longinput=long_med, timeinput=time_sq)})
stopCluster(cl)
###
library(tidyr)
anglong <- bind_rows(list_angs)
angwide <- spread(anglong, key=eq_angle_by_10,value=total)
rm(anglong, list_angs, time_sq)
save(angwide, file="../othereqdata/eq_australia_expected.RData")
}
## ------------------------------------------------------------------------
load("../othereqdata/eq_australia_processed.RData")
load("../othereqdata/eq_australia_expected.RData")
eq_night = sum(eq_national$eq_is_night)
eq_total = nrow(eq_national)
bands <- rev(c('#ffffcc','#d9f0a3','#addd8e','#78c679','#41ab5d','#238443','#005a32'))
sigmas <- c(0.682689492137086,
0.954499736103642,
0.997300203936740,
0.999936657516334,
0.999999426696856,
0.999999998026825,
0.999999999997440)
lbls <- c(
"1 sigma", "2 sigma",
"3 sigma", "4 sigma",
"5 sigma", "6 sigma",
"7 sigma")
typs <- c(1,1,1,1,1,1,1)
weights <- c(3,3,3,3,3,3,3)
old_par=par()
## ------------------------------------------------------------------------
bt <- binom.test(eq_night ,eq_total, conf.level= .999999999997440)
## ------------------------------------------------------------------------
feature <- c("Earliest (UTC)", "Latest (UTC)",
"Northernmost", "Southernmost",
"Westmost", "Eastmost",
"Percent < Mag 3", "total entries",
"nighttime quakes")
value <- c(as.character(min(eq_national$time_UTC)),
as.character(max(eq_national$time_UTC)),
as.character(max(eq_national$latitude)),
as.character(min(eq_national$latitude)),
as.character(min(eq_national$longitude)),
as.character(max(eq_national$longitude)),
as.character(round(100*sum(eq_national$magnitude < 3)/eq_total,2)),
as.character(eq_total),
as.character(eq_night))
data.frame(feature,value) %>% knitr::kable(caption="Data description")
## ---- fig.cap="Proportion of earthquakes at night: Philippines. n=8054"----
### making the basic proportion graph
eq_night = sum(eq_national$eq_is_night)
eq_total = nrow(eq_national)
bands <- rev(c('#ffffcc','#d9f0a3','#addd8e','#78c679','#41ab5d','#238443','#005a32'))
sigmas <- c(0.682689492137086,
0.954499736103642,
0.997300203936740,
0.999936657516334,
0.999999426696856,
0.999999998026825,
0.999999999997440)
lbls <- c(
"1 sigma", "2 sigma",
"3 sigma", "4 sigma",
"5 sigma", "6 sigma",
"7 sigma")
typs <- c(1,1,1,1,1,1,1)
weights <- c(3,3,3,3,3,3,3)
old_par=par()
conf_steps <- function(x, sigmas=sigmas, night=eq_night, total=eq_total){
ci_lower <- binom.confint(night, total, method=c("wilson"), conf.level = sigmas[x])[1,5]
ci_upper <- binom.confint(night, total, method=c("wilson"), conf.level = sigmas[x])[1,6]
ci_data <- data.frame(step = x, ci_lower, ci_upper)
}
ci_spacing <- lapply(7:1, conf_steps, sigmas=sigmas, night=eq_night, total=eq_total)
ci_steps <- bind_rows(ci_spacing)
layout(matrix(c(1,1,1,2), ncol=4))
par(mar=c(5,6,4,2))
plot(c(min(0.5,floor(100*ci_steps[1,2])/100), max(0.5,ceiling(100*ci_steps[1,3])/100)),
y=c(-3,8), type="n", bty="n", yaxt="n", ylab="",
xlab="Proportion of earthquakes at night")
a <- a <- apply(ci_steps, 1, function(x){
polygon(c(x[2], x[3], x[3], x[2]), c(0, 0, 1, 1), col=bands[x[1]], border=NA)})
lines(c(.5,.5), c(0,1), col="#FFFFFF")
lines(c(.5,.5), c(0,1), lty=2, col="#777777")
lines(c(eq_night/eq_total,eq_night/eq_total), c(0,1), lwd=2)
par(mar=c(0,0,0,0))
plot(x=c(0,10), y=c(0,10), type="n", bty="n", axes=FALSE)
legend(0,5.5, legend=lbls, lty=typs, lwd=weights, col=bands, bty="n", xjust=0,
title="Confidence Intervals:", y.intersp=1.1, cex=0.9)
lbls2=c("50% Night", "Actual Proportion")
typs2=c(2,1)
weights2=c(1,2)
cls2=c("#777777","#000000")
legend(0,7, legend=lbls2, lty=typs2, lwd=weights2, col=cls2, bty="n", xjust=0,
title="Legend", y.intersp=1.2)
par(mar=old_par$mar)
par(mfrow=c(1,1))
## ---- fig.cap="Proportion of night earthquakes by magnitude, Philippines. n=8054"----
old_par=par()
grf <- eq_national %>% mutate(floored_mag = floor(magnitude*2)/2) %>%
group_by(floored_mag) %>% summarise(successes = sum(eq_is_night), trials=n())
poly_conf_int <- function(success, trials, aa, stepsize, sigma, colr){
ci <- binom.confint(success, trials, method=c("wilson"), conf.level = sigma)
lower <- ci[1,5]
upper <- ci[1,6]
a <- polygon(x=c(aa,aa+stepsize,aa+stepsize,aa), y=c(upper,upper,lower,lower),
col=colr, border=NA)
}
plot7sig <- function(success, trials, aa, stepsize){
library(binom)
#bands <- c('#ffffb2','#fed976','#feb24c','#fd8d3c','#fc4e2a','#e31a1c','#b10026')
bands <- rev(c('#ffffcc','#d9f0a3','#addd8e','#78c679','#41ab5d','#238443','#005a32'))
sigmas <- c(0.682689492137086,
0.954499736103642,
0.997300203936740,
0.999936657516334,
0.999999426696856,
0.999999998026825,
0.999999999997440)
sapply(7:1, function(x){
poly_conf_int(success, trials, aa, stepsize, sigmas[x], bands[x])})
a <- lines(c(aa, aa + stepsize), c(success/trials, success/trials), lwd=2)
}
lbls <- c(
"1 sigma", "2 sigma",
"3 sigma", "4 sigma",
"5 sigma", "6 sigma",
"7 sigma")
typs <- c(1,1,1,1,1,1,1)
weights <- c(3,3,3,3,3,3,3)
clrs <- rev(c('#ffffcc','#d9f0a3','#addd8e','#78c679','#41ab5d','#238443','#005a32'))
#clrs <- c('#ffffb2','#fed976','#feb24c','#fd8d3c','#fc4e2a','#e31a1c','#b10026')
layout(matrix(c(1,1,1,2), ncol=4))
plot(x=c(0,max(grf$floored_mag)+0.5), y=c(0,1), type="n", bty="n",
xlab="Magnitude (0.5 steps)", ylab="Proportion of earthquakes at night")
a <- apply(grf,1,function(x){plot7sig(x[2],x[3],x[1],0.5)})
lines(c(0,10), c(.5,.5), col="#FFFFFF")
lines(c(0,10), c(.5,.5), lty=2, col="#777777")
par(mar=c(0,0,0,0))
plot(x=c(0,10), y=c(0,10), type="n", bty="n", axes=FALSE)
legend(0,5, legend=lbls, lty=typs, lwd=weights, col=clrs, bty="n", xjust=0,
title="Confidence
Intervals:", cex=0.9)
lbls=c("Expected Proportion", "Actual Proportion")
typs=c(2,1)
weights=c(1,2)
legend(0,7, legend=lbls, lty=typs, lwd=weights, bty="n", xjust=0,
title="Legend", y.intersp=1.2)
par(mar=old_par$mar)
par(mfrow=c(1,1))
## ------------------------------------------------------------------------
by_angle <- eq_national %>%
group_by(eq_angle_by_10) %>% summarise(total= n()) %>%
mutate(daynight=ifelse(eq_angle_by_10 < 180, "day", "night"))
merged <- merge(eq_national, angwide, by.x="eq_roundedlat", by.y="lat")
agg_expected <- merged %>% select(`0`:`350`) %>% colSums(na.rm=TRUE)
expected_prop <- agg_expected / sum(agg_expected)
expected <- data.frame(eq_angle_by_10 = as.numeric(names(expected_prop)),
expected_prop = as.numeric(expected_prop))
expected$expected_number = expected_prop * eq_total
act_exp <- merge(expected, by_angle, by="eq_angle_by_10", all.x=TRUE)
act_exp$total[is.na(act_exp$total)] <- 0
act_exp$daynight <- NULL
act_exp$act_prop <- act_exp$total / sum(act_exp$total)
ci_brackets <- act_exp %>% ungroup() %>% mutate(grand_total=sum(total)) %>%
rowwise() %>% mutate(
ci_lower_7 = binom.confint(total, grand_total, method=c("wilson"),
conf.level = sigmas[7])[1,5] * grand_total,
ci_upper_7 = binom.confint(total, grand_total, method=c("wilson"),
conf.level = sigmas[7])[1,6] * grand_total,
ci_lower_6 = binom.confint(total, grand_total, method=c("wilson"),
conf.level = sigmas[6])[1,5] * grand_total,
ci_upper_6 = binom.confint(total, grand_total, method=c("wilson"),
conf.level = sigmas[6])[1,6] * grand_total,
ci_lower_5 = binom.confint(total, grand_total, method=c("wilson"),
conf.level = sigmas[5])[1,5] * grand_total,
ci_upper_5 = binom.confint(total, grand_total, method=c("wilson"),
conf.level = sigmas[5])[1,6] * grand_total,
ci_lower_4 = binom.confint(total, grand_total, method=c("wilson"),
conf.level = sigmas[4])[1,5] * grand_total,
ci_upper_4 = binom.confint(total, grand_total, method=c("wilson"),
conf.level = sigmas[4])[1,6] * grand_total,
ci_lower_3 = binom.confint(total, grand_total, method=c("wilson"),
conf.level = sigmas[3])[1,5] * grand_total,
ci_upper_3 = binom.confint(total, grand_total, method=c("wilson"),
conf.level = sigmas[3])[1,6] * grand_total,
ci_lower_2 = binom.confint(total, grand_total, method=c("wilson"),
conf.level = sigmas[2])[1,5] * grand_total,
ci_upper_2 = binom.confint(total, grand_total, method=c("wilson"),
conf.level = sigmas[2])[1,6] * grand_total,
ci_lower_1 = binom.confint(total, grand_total, method=c("wilson"),
conf.level = sigmas[1])[1,5] * grand_total,
ci_upper_1 = binom.confint(total, grand_total, method=c("wilson"),
conf.level = sigmas[1])[1,6] * grand_total)
norm_ci <- ci_brackets
for (i in c(4,7:20)){
norm_ci[,i] <- ci_brackets[,i] - ci_brackets[,3]
}
circlesize=100
## ---- fig.cap="Over- and under- supply of earthquakes by angle of the sun
## (10 degree steps). Hawaii. n=10181"----
norm_ci$border = 2
# need to double entries with a displacement of 10
# to make each side of the item on the graph
norm_ci2 <- norm_ci
norm_ci2$eq_angle_by_10 <- norm_ci2$eq_angle_by_10 + 10
norm_ci2$border = 1
graphdata <- bind_rows(norm_ci,norm_ci2) %>% arrange(eq_angle_by_10,border)
#### plot graph
bands <- rev(c('#ffffcc','#d9f0a3','#addd8e','#78c679','#41ab5d','#238443','#005a32'))
old_par=par()
layout(matrix(c(1,1,1,2), ncol=4))
# overall limits
limits=2 * max(abs(c(graphdata$ci_lower_7, graphdata$ci_upper_7)))
# plot upper confidence 7 interval using plotrix
polar.plot(graphdata$ci_upper_7, polar.pos=graphdata$eq_angle_by_10,
radial.lim=c(-1*limits,limits),
labels = "", main=NULL,lwd=0.5, rp.type="p",
show.grid.labels=FALSE, show.grid=FALSE, mar=c(0,0,0,0),
grid.col=bands[7], line.col=bands[7], poly.col=bands[7])
# plot upper 6 confidence interval
plot_ci_round <- function(upper_bound,x){
polar.plot(upper_bound, polar.pos=graphdata$eq_angle_by_10, add=TRUE,
radial.lim=c(-1*limits,limits),
line.col=bands[x], lwd=0.5, rp.type="p", poly.col=bands[x])
}
plot_ci_round(graphdata$ci_upper_6, 6)
plot_ci_round(graphdata$ci_upper_5, 5)
plot_ci_round(graphdata$ci_upper_4, 4)
plot_ci_round(graphdata$ci_upper_3, 3)
plot_ci_round(graphdata$ci_upper_2, 2)
plot_ci_round(graphdata$ci_upper_1, 1)
plot_ci_round(graphdata$ci_lower_1, 2)
plot_ci_round(graphdata$ci_lower_2, 3)
plot_ci_round(graphdata$ci_lower_3, 4)
plot_ci_round(graphdata$ci_lower_4, 5)
plot_ci_round(graphdata$ci_lower_5, 6)
plot_ci_round(graphdata$ci_lower_6, 7)
polar.plot(graphdata$ci_lower_7, polar.pos=graphdata$eq_angle_by_10,
add=TRUE, radial.lim=c(-1*limits,limits),
line.col="white", lwd=0.5, rp.type="p", poly.col="white")
# plot expected guide line
polar.plot(rep(0,nrow(graphdata)), polar.pos=graphdata$eq_angle_by_10, add=TRUE,
radial.lim=c(-1*limits,limits),
rp.type="p", lty=4)
# plot 500 less than expected guide line
polar.plot(rep(-1 * circlesize,nrow(graphdata)), polar.pos=graphdata$eq_angle_by_10,
add=TRUE,radial.lim=c(-1*limits,limits),
rp.type="p", lty=1, line.col="#00000044")
# plot 500 more than expected guide line
polar.plot(rep(circlesize,nrow(graphdata)), polar.pos=graphdata$eq_angle_by_10,
add=TRUE,radial.lim=c(-1*limits,limits),
rp.type="p", lty=3, line.col="#00000044")
lines(c(-1.5,-1.2)*limits, c(0,0))
lines(c(1.5,1.2)*limits, c(0,0))
text(-1.8*limits,0, label="sunset
180", cex=0.7)
text(1.8*limits,0, label="sunrise
0", cex=0.7)
par(mar=c(0,0,0,0))
plot(x=c(0,10), y=c(0,10), type="n", bty="n", axes=FALSE, xlab="")
lbls <- c(
"1 sigma", "2 sigma",
"3 sigma", "4 sigma",
"5 sigma", "6 sigma",
"7 sigma")
typs <- c(1,1,1,1,1,1,1)
weights <- c(3,3,3,3,3,3,3)
clrs <- rev(c('#ffffcc','#d9f0a3','#addd8e','#78c679','#41ab5d','#238443','#005a32'))
legend(0,4.5, legend=lbls, lty=typs, lwd=weights, col=clrs, bty="n", xjust=0,
title="Confidence Intervals:", cex=0.9)
lbls2=c("Expected Number", paste(circlesize,"under expected"),
paste(circlesize,"over expected"))
typs2=c(4,1,3)
weights2=c(1,1,1)
clrs2=c("#000000","#00000044","#00000044")
legend(0,10, legend=lbls2, lty=typs2, lwd=weights2, bty="n", xjust=0,
title="Legend", y.intersp=1.2, col=clrs2)
par(mar=old_par$mar)
par(mfrow=c(1,1))